Viewpoint selection for molecular visualization: analysis and applications
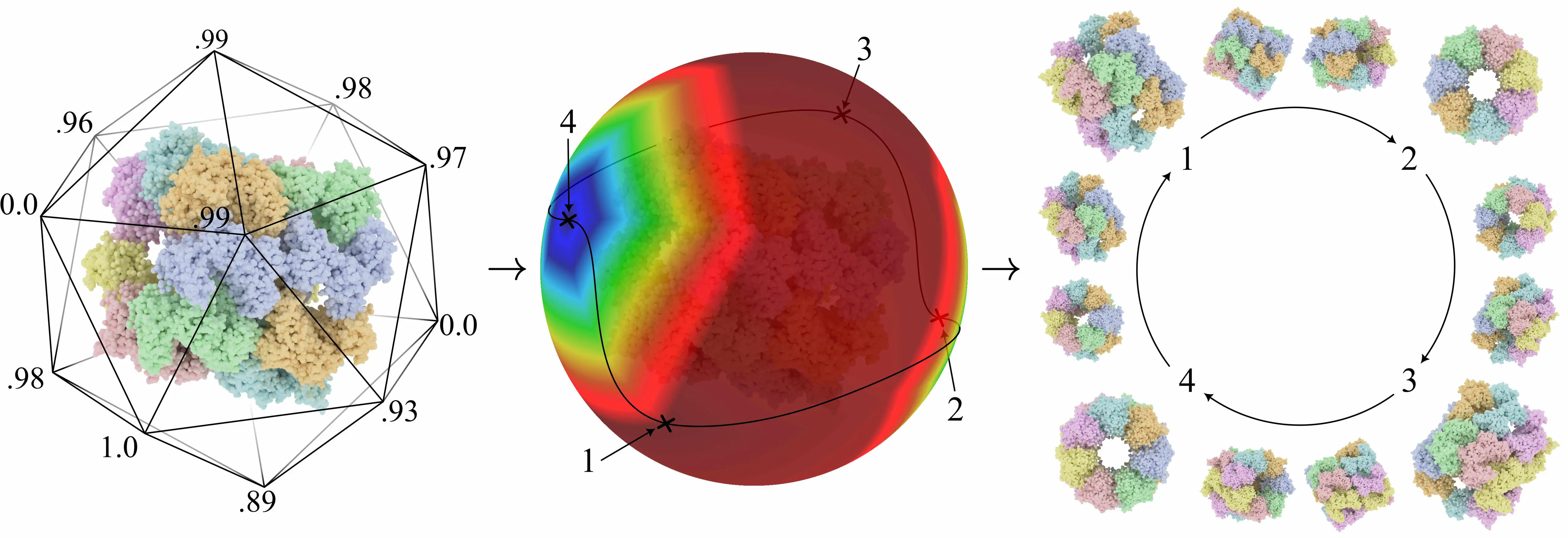 Tour creation pipeline for a molecule. From viewpoint selection to camera animation.
Tour creation pipeline for a molecule. From viewpoint selection to camera animation.Abstract
Molecular systems are often visually complex and abstract. It can be difficult, even for experts, to explore and find meaningful viewpoints. Research in viewpoint selection methods has mainly focused on general real-world objects and only a few works tackled molecular scenes. In this paper, we present a study of 20 state-of-the-art viewpoint selection methods from the general field, applied to molecular visualization. Our goal is to determine if these methods can find two important geometrical configurations for molecules. Additionally, we propose an automatic generation of informative and visually pleasing molecular tours to help the study and the communication around molecules.
Viewpoint selection
Viewpoint selection is done using metrics, which are functions $ f(v) = s $ which give a score $ s $ from the image of a viewpoint $ v $.
We studied 20 metrics divided in 5 categories from previous works1 2 for finding two geometrical configurations of molecules: the alignement and the tunnel.
| Alignement3 | Tunnel4 |
|---|---|
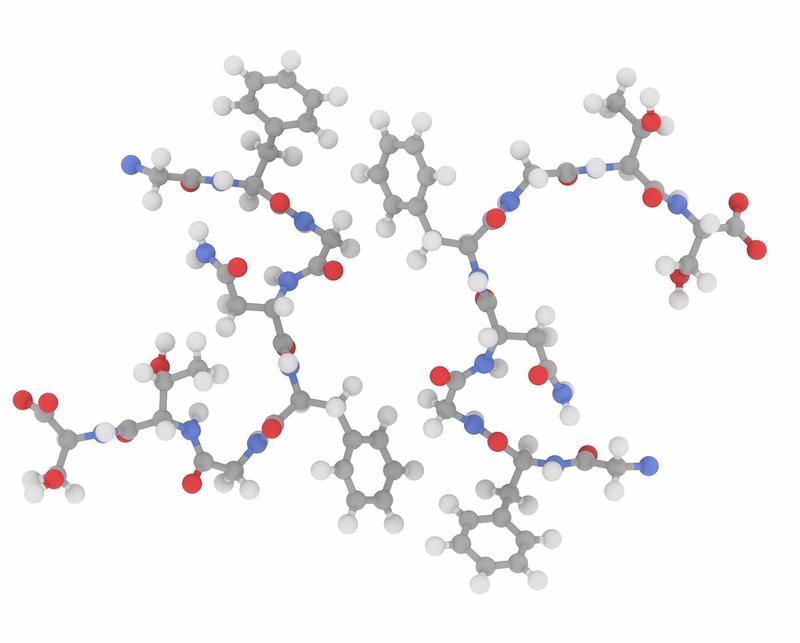 |  |
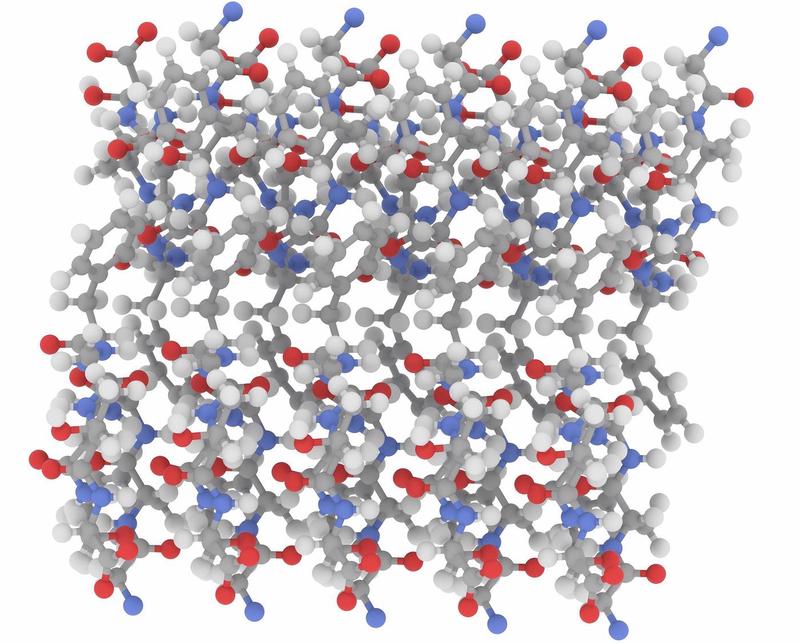 | 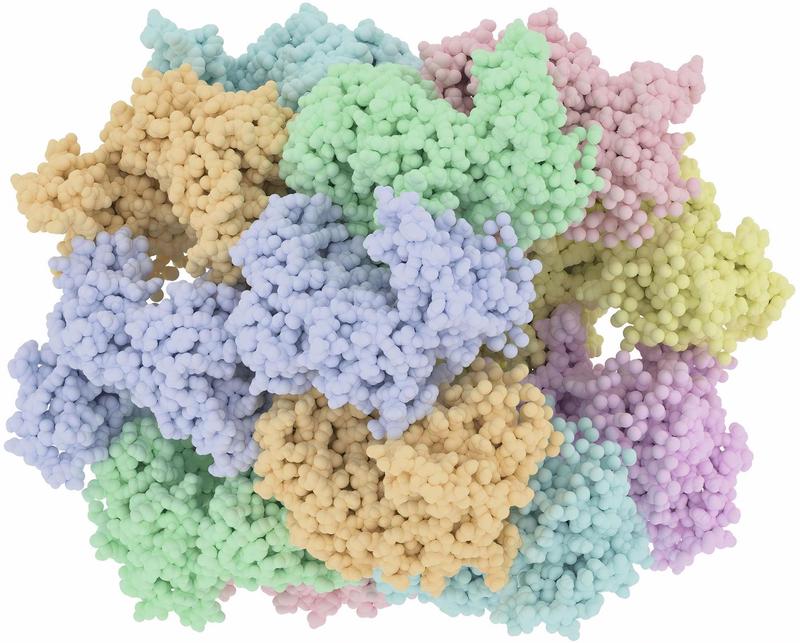 |
Top: the geometrical configuration. Bottom: another viewpoint of the same molecule.
Each metric is evaluated on its:
- Accuracy: find a configuration either on highest or lowest score.
- Consistency: find a configuration only on highest or lowest score.
Metrics results for the alignment
Metrics results for the tunnelMolecular tours
To create the molecular tour we select viewpoints around the molecule so that they are spread around the molecule as much as possible. We separatly select viewpoint with high and low scores and connect them using a Catmull Rom spline to have a smooth path. Finally we animate the camera to respect some animation principle which gives the following result:
Exemple molecular tour with 4 key viewpoints.
Paper citation:Bibtex
@conference{LMMM24,
author={Vincent Larroque. and Maxime Maria. and Stéphane Mérillou. and Matthieu Montes.},
title={Viewpoint Selection for Molecular Visualization: Analysis and Applications},
booktitle={Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications - GRAPP},
year={2024},
pages={58-69},
publisher={SciTePress},
organization={INSTICC},
doi={10.5220/0012383300003660},
isbn={978-989-758-679-8},
issn={2184-4321},
}
Harvard
Larroque, V.; Maria, M.; Mérillou, S. and Montes, M. (2024). Viewpoint Selection for Molecular Visualization: Analysis and Applications. In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications - GRAPP; ISBN 978-989-758-679-8; ISSN 2184-4321, SciTePress, pages 58-69. DOI: 10.5220/0012383300003660
Secord et al. - Perceptual Models of Viewpoint Preference - ACM Transactions on Graphics - 2011 ↩︎
Bonaventura et al. - A Survey of Viewpoint Selection Methods for Polygonal Models - Entropy - 2018 ↩︎
Vázquez et al. - Viewpoint entropy: a new tool for obtaining good views of molecules - Proceedings of the symposium on Data Visualisation - 2002 ↩︎
Heinrich et al. - Evaluating Viewpoint Entropy for Ribbon Representation of Protein Structure - Computer Graphics Forum - 2016 ↩︎