Improving Consistency of Viewpoint Selection for Molecules
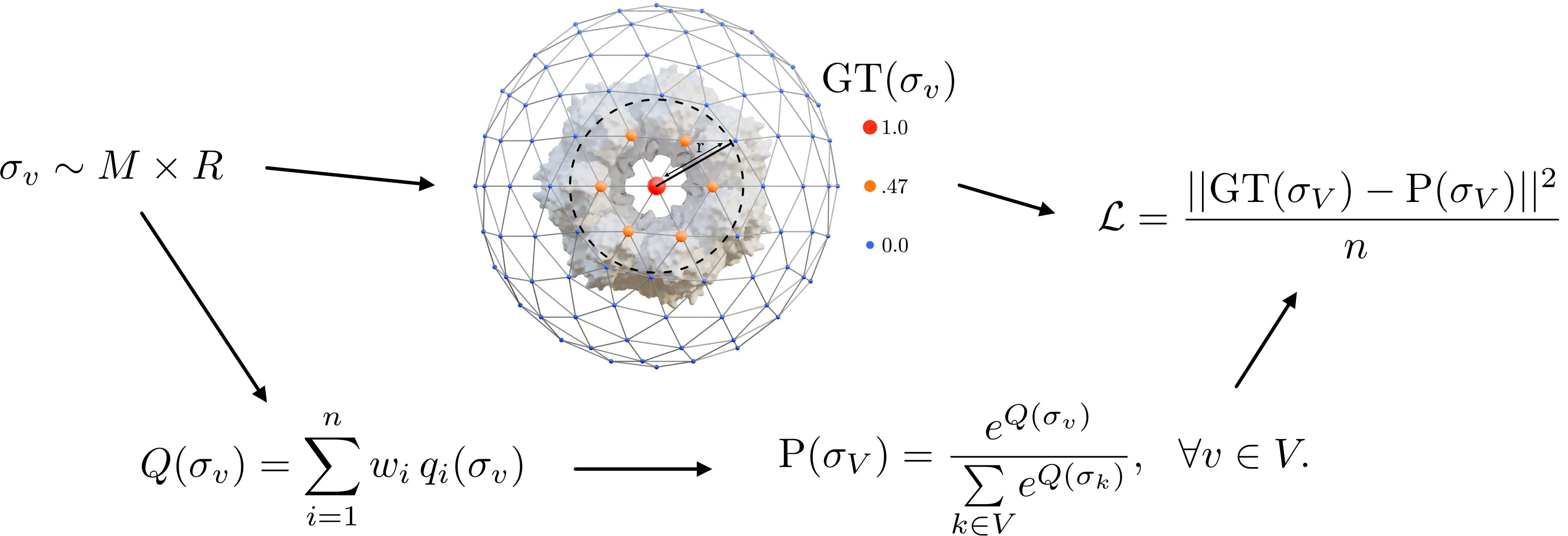 Illustrative pipeline showing the construction of the loss function from a scene configuration.
Illustrative pipeline showing the construction of the loss function from a scene configuration.Abstract
Molecular visualization is widely used in structural molecular biology and drug discovery to study the structure of molecules and help their understanding. However, identifying meaningful viewpoints is challenging because of their abstract and convoluted nature of molecular scenes. Consequently, viewpoint selection methods have been developed to highlight important features of molecules. These methods base the selection process on metrics, which are functions that assign a score based on what can be observed in an image of a molecule. In this paper, we study 20 state-of-the-art metrics across four common molecular representations: Ball & Stick, van der Waals, Solvent Excluded Surface, and Cartoon. We evaluate the consistency of these metrics in identifying two crucial geometrical configurations for molecular studies: alignment and tunnel. We show that these metrics do not provide consistent results in finding these features. Therefore, we propose to optimize linear metric combination models, which significantly improves the overall detection performance.